Example: 2D Correlation and Prototype Matching
Illustrates the correlation of a 2D kernel with an image in the spatial domain.
One interesting kernel is the Gaussian kernel, which produces a smoothing effect.
For information on using this example, refer to About Image Processing Examples.
2D Correlation
1. Use the READ_IMAGE function to read in an image.
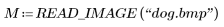
2. Use the WRITEBMP function to save the image into a file.
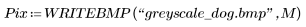
3. Define the kernel range and function.
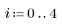
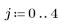
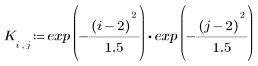
4. Evaluate the 5 x 5 kernel matrix.
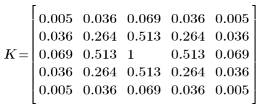
This kernel is symmetric, so the effect is the same as convolution - equivalent to correlation with the kernel reversed.
5. Define a scale factor mcscale.
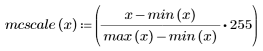
6. Calculate the cross correlation, then scale the result to 8-bit grayscale from 0 to 255.
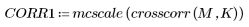
7. Use the WRITEBMP function to save the results into a file.
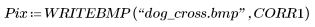
8. Plot the original image and the correlation of the image with the kernel.
 (greyscale_dog.bmp) |  (dog_cross.bmp) |
Prototype Matching
1. Use the READ_IMAGE function to read in a new image.
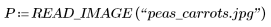
2. Use the WRITEBMP function to save the results into a file.
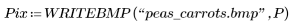

(peas_carrots.bmp)
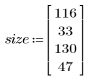
The selection picks out a representative carrot as a template.
3. Define the height and width of the template.
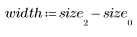
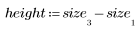
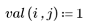
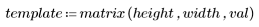
5. Plot the template and the carrot images.
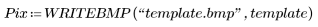
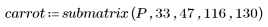
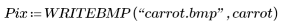
 (template.bmp) |  (carrot.bmp) |
6. Define the correlation threshold.
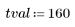
7. Use the uniform template of the same size as the selected carrot, or use the selected representative carrot image itself to calculate the number of matches:
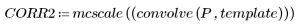
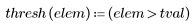
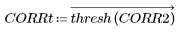
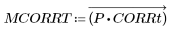
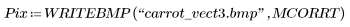
 (carrot_vect.bmp) |
Most of the peas are blacked out by the uncorrelated, threshold values, and most of the carrots are still visible. The match is made between the original image and the rectangular template of uniform density, which roughly picks out the carrots in this random sampling from a frozen vegetable package.
8. Calculate the approximate percentage of carrots in the original image.
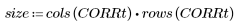
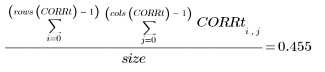
This technique is also useful for bacteria counting, and other image feature percentage estimation.